Researchers at Washington University School of Medicine in St. Louis have revealed the structure of a key protein involved in unspooling DNA so that it can undergo repair. Because this protein is critical for DNA repair for many organisms, including those that cause diseases such as M. tuberculosis and E. coli, understanding how it operates could lead to new ways of blocking the growth of pathogens.
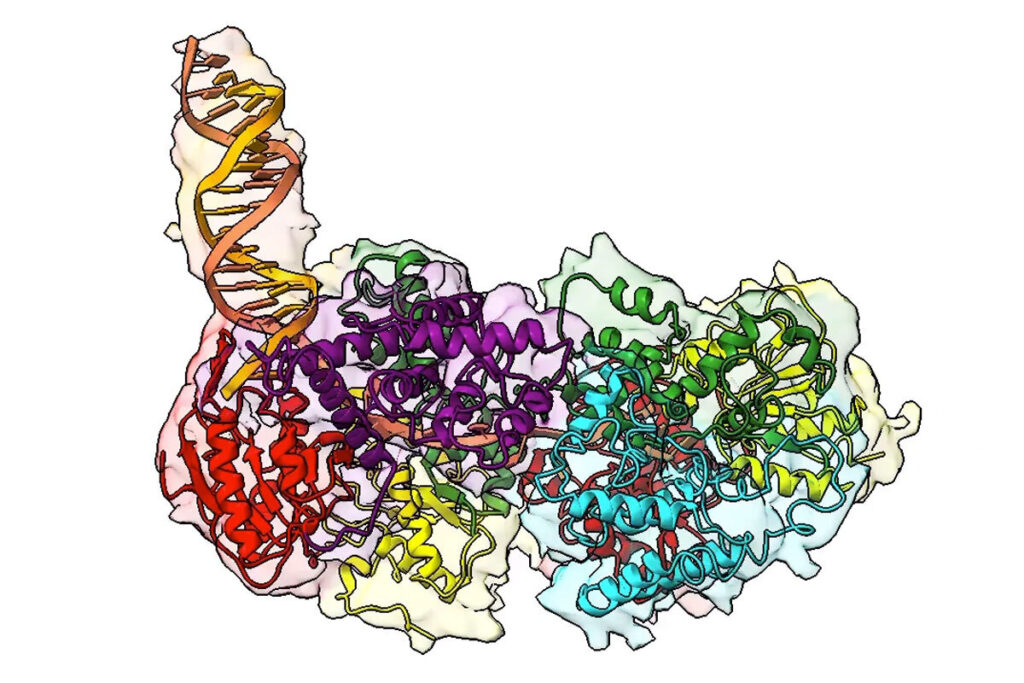
For many years, biochemical data suggested that this protein — a helicase enzyme called UvrD1 — must be assembled from two smaller subunits to be able to unzip the DNA molecule. But structural analyses only ever revealed a singular complex, known as a monomer. Now, a recent study in the Proceedings of the National Academy of Sciences reveals the first two-part structure — known as a dimer — of this key helicase enzyme, answering questions about the 3D structure and regulation of the enzyme activity that have dogged the field for decades.
The study was led by Eric Galburt, a professor of biochemistry and molecular biophysics; and Ankita Chadda, who conducted this work as a graduate student in the Galburt lab and is now a postdoctoral researcher at the Salk Institute for Biological Studies in San Diego; along with Timothy Lohman, the Marvin A. Brennecke Professor of Biophysics, and Binh Nguyen, a staff scientist in the Lohman lab.
Their research also suggests possible ways to interfere with the UvrD1 enzyme, which could prove useful in developing new therapeutics for tuberculosis or other diseases caused by organisms that rely on these helicases to repair their DNA.