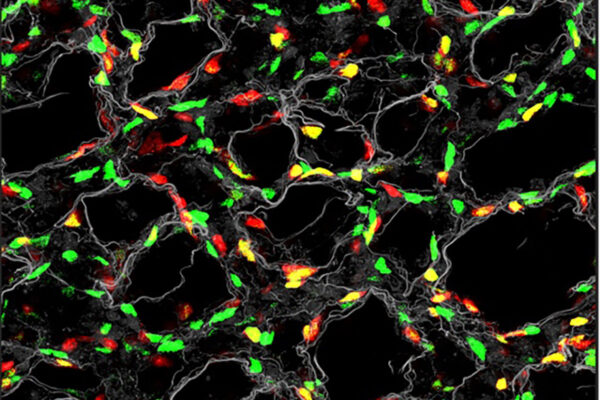
Most well-studied proteins are folded, meaning they have a defined three-dimensional shape that helps determine each protein’s specific function. But as the tools of science have improved, so has the understanding that many important proteins — or sections of proteins — don’t maintain a fixed shape even as they carry out vital cellular processes.
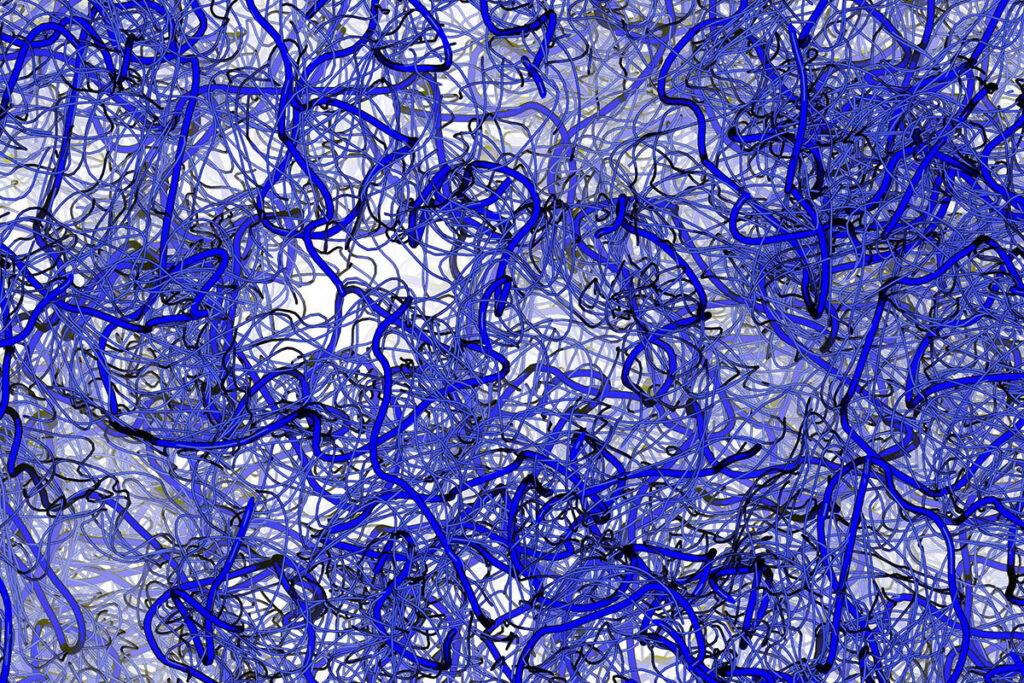
Called intrinsically disordered proteins, these wiggly structures are in constant motion and, as a result, are difficult to study. While computational tools such as AlphaFold — which won the 2024 Nobel Prize in Chemistry — have been transformative for understanding how folded proteins work, AlphaFold does not work well for studying these wiggly proteins. A better understanding of these shape-shifting proteins could identify previously unknown causes of human disease and present novel approaches to new therapies.
Now, research published in the journal Science and led by WashU Medicine’s Alex Holehouse, an assistant professor of biochemistry and molecular biophysics, describes a computational method his team developed to help predict how intrinsically disordered proteins will behave. The tool analyzes the chemical interactions of the proteins’ building blocks, called amino acids, and predicts which bits of the disordered protein will be attractive or repulsive for other molecules in the body. By predicting based on chemistry alone, the approach allows scientists to model interactions even in the absence of a defined 3D structure.
The method — called FINCHES, an acronym for First-principle INteractions via CHEmical Specificity — can help develop molecular hypotheses about protein activities and interactions that can then be tested in the lab. This means researchers can begin to quickly explore how disordered proteins might work in many critical biological contexts and, importantly, how mutations might disrupt protein function in the context of human disease, notably cancer and neurodegeneration. FINCHES is open source and available to investigators as a Python package and via a web server.