In the world of molecular biology, getting high-quality data from tiny biological systems while they’re in motion is something like trying to take a clear picture of a spinning propeller. Researchers need advanced techniques and careful calculations to measure such movement accurately.
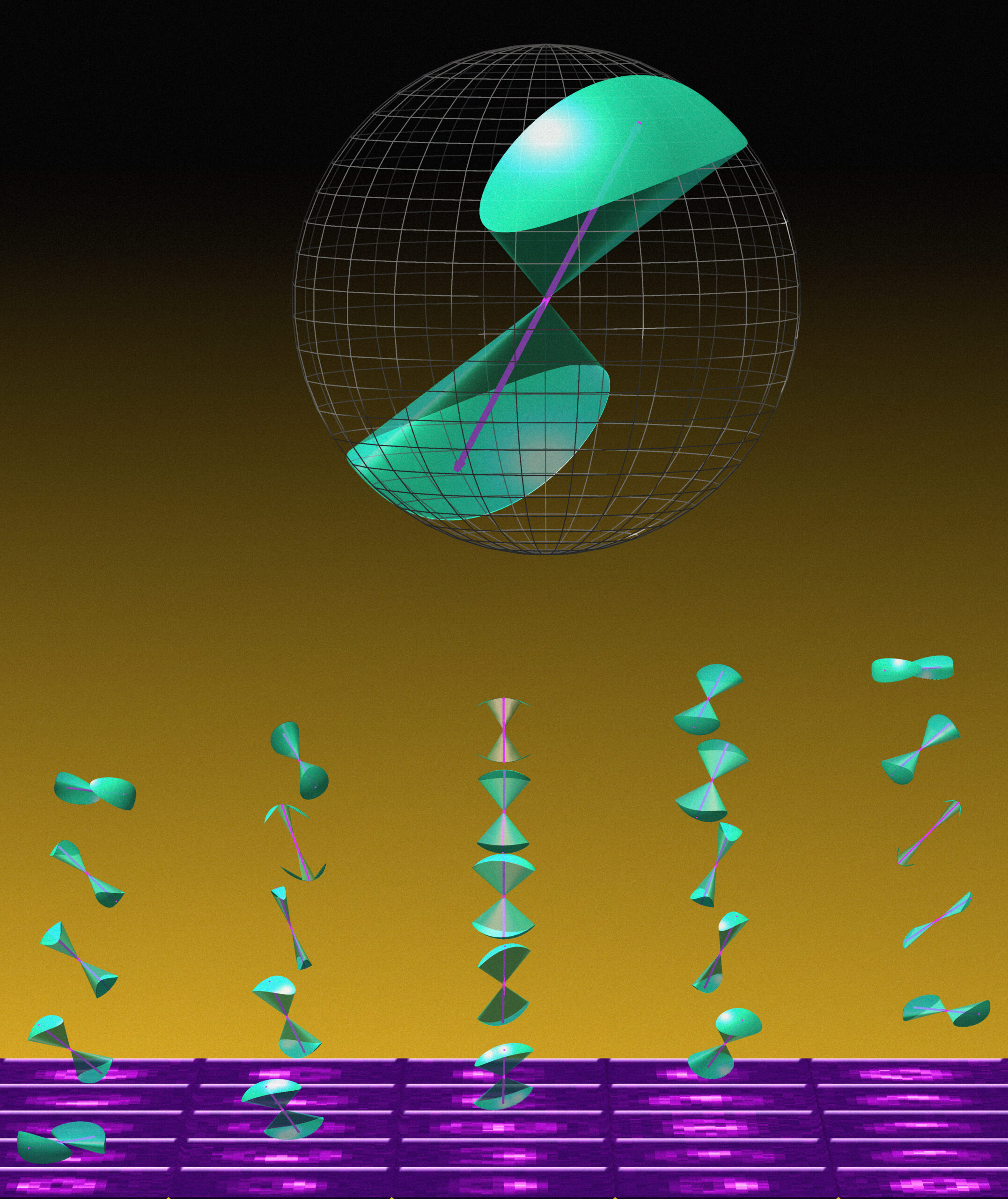
Traditionally, these methods have relied on simplifying assumptions that overlook some complexities of molecular behavior, which can be wobbly and asymmetric. A new theoretical framework developed by Washington University in St. Louis’ Matthew Lew, however, is set to shake up such measurements.
In a cover article published July 8 in the Journal of Physical Chemistry A, Weiyan Zhou, a PhD student in electrical engineering, and Lew introduced a detailed model that allows scientists to more accurately describe and measure how molecules move.
Where traditional techniques assume that molecules wobble uniformly in all directions within a circular cone, Lew, an associate professor of electrical and systems engineering at the McKelvey School of Engineering, discarded this approach to reflect the true nature of molecular behavior in more complex biological environments.
This increased precision will be especially valuable in applications such as immunology or the study of biomolecular condensates.
Read more on the McKelvey Engineering website.