Gary Patti, the Michael and Tana Powell Associate Professor of Chemistry in Arts & Sciences at Washington University in St. Louis, has been awarded $4.8 million in two separate National Institutes of Health (NIH) grants focused on improving the accessibility of metabolomics — the study of the biochemical reactions that underlie metabolism.
Just as the genome is a complete set of the genes or genetic material present in a cell or organism, the metabolome represents a complete set of the metabolites — such as sugars, amino acids, fats and other more unfamiliar small molecules — in a cell or organism under a particular set of conditions.
Researchers such as Patti seek to create comprehensive metabolic profiles that can be compared between healthy and disease states.
“As it turns out, metabolic profiles are highly informative with respect to human health,” Patti said. Metabolites are already used in clinics around the world to diagnose many different diseases, such as cancer. But current tests used in hospitals look at a relatively small number of metabolites compared to the hundreds to thousands of signals observed in metabolomics. “There’s a big opportunity here not only to move to better diagnostics but also to better understand the fundamental biochemistry of disease,” he said.
Finding what feeds cancer
There are trillions of cells in the human body. Patti is interested in how metabolism varies between them.
“In many ways, metabolic processes are a product of cellular environment,” Patti said. “A person living on the coast might be more likely to eat seafood; whereas, we are more likely to eat toasted ravioli here in St. Louis. Cells are the same way.”
Patti believes that tumors have their own unique “neighborhoods” and nutritional preferences. If he can determine which metabolites “feed” cancer cells, then perhaps there are ways to limit cancer’s access to key ingredients in the body that can stop or limit cancer cells’ growth.
That’s where modern metabolomics comes into play. The tools that form the backbone of metabolomics have been around for more than 15 years. But because of the complexity of data interpretation — which requires knowledge of mass spectrometry, statistics and multiple computer programming languages — only a small percentage of related studies that are published each year rely on the kind of cutting-edge metabolomics technologies that NIH has invested in with its six research centers, or Regional Comprehensive Metabolomics Resource Cores.
All that could change if there was a standard, accepted way to convert the large number of signals returned by metabolomics tests to a simple list corresponding to the actual metabolites present.
“When you do metabolomics, you often detect thousands of signals,” Patti said. “We and others have done work to show that there are many more signals than metabolites. The question is: How can we decipher metabolites from all of the other junk? Biologists and clinicians only want to know what metabolites are there and what their concentrations are. That’s the gap.”
The signals that matter
The process is arduous, but getting rid of noise signals is important. The majority of signals detected — up to 95 percent in some experiments, Patti said — correspond not to intact metabolites, but instead to contamination, artifacts, and other signals that are difficult to interpret. For example, a metabolite may break apart into multiple pieces, each producing its own signal.
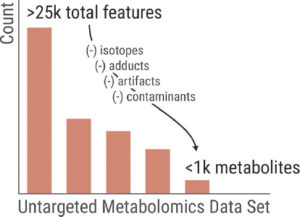
A component of one of Patti’s new grants is focused on separating out the signals that matter and developing comprehensive lists that other researchers can reference. For this $1.8 million, four-year program, Patti’s group at Washington University is one of seven laboratory teams each supplying separate expertise to a collaborative effort led by the National Cancer Institute at the NIH. The program is supported by a U-grant mechanism, which represents a partnership between the research groups and the NIH.
The other new grant is a $3 million, four-year effort focused on the metabolism of C. elegans worms and zebrafish.
“Of course, improving human health is the ultimate goal. But we can get a lot of insight into the biochemistry of humans by studying model organisms such as worms and zebrafish,” Patti said. Whether a worm, fish or human, development from an embryo to an adult requires a lot of growth. Patti aims to figure out which metabolic processes are essential to each step of the process.
“We can then compare the metabolic profiles of growing healthy tissue to growing cancer tissue,” he said. “We are always looking for metabolic differences between healthy and disease states, because these differences represent potential therapeutic opportunities.”
Taming the output
Patti’s work in metabolomics takes a page from Washington University’s rich history in genomics.
“We’re applying very similar types of strategies to study metabolism with metabolomics that have proven to be successful in genomics,” he said. “Taming the output of genome sequencing experiments not only extended their reach, it also improved throughput.”
More throughput means more studies with the best modern tools — without the data burden.
“Your everyday biologist or clinician who is interested in metabolism may not have access to all of the computational areas of expertise needed for metabolomics,” Patti said. “We’re focused on trying to lower that barrier, and alleviate some of that data burden — so that a physician who isn’t trained extensively in this field can come and do the experiment, get a result, and say ‘I know what I can do with this.’”