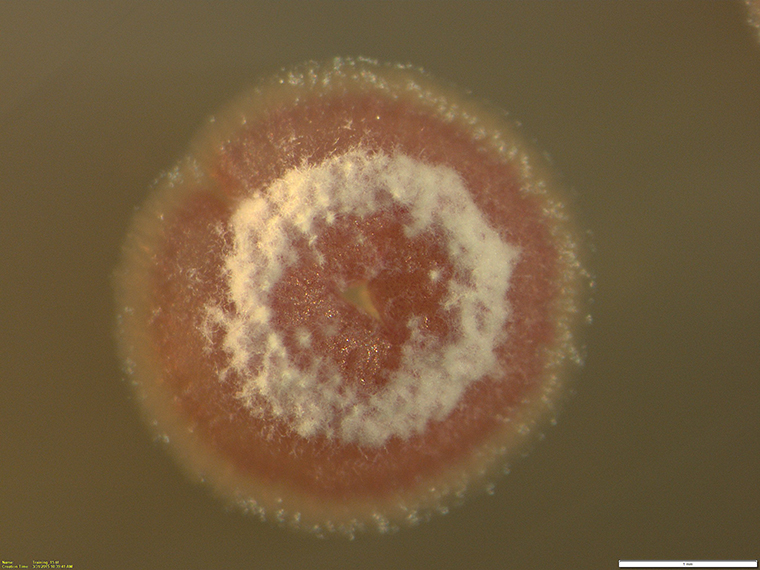
An untapped trove of desirable drug-like molecules is hidden in the genomes of Streptomyces bacteria — the same bacteria responsible for the first bacterial antibiotics to treat tuberculosis back in the 1940s.
Isolating them, however, has proved challenging. Now, biologists at Washington University in St. Louis are using comparative metabologenomics to try to uncover what may be “silencing” Streptomyces and preventing it from producing desirable compounds encoded by its genes.

“We examined genetic differences across the genomes of Streptomyces while at the same time looking at antibiotic outputs,” said Joshua Blodgett, assistant professor of biology in Arts & Sciences, the corresponding author of research published this week in the Proceedings of the National Academy of Sciences (PNAS). “This study highlights comparative metabologenomics as a powerful approach to expose the features that differentiate strong antibiotic producers from weaker ones.”
Blodgett’s team, including recent PhD graduate Yunci Qi and postdoctoral research associate Keshav Nepal, compared a group of antibiotic-producing strains of Streptomyces and other nonproducing or poor-producing strains to reveal genomic differences that could affect drug production.
The researchers found a few key differences between the strains. Notably, the good producers of polycyclic tetramate macrolactam (PTM) antibiotics seemed to benefit from griseorhodin production, which the researchers did not anticipate and originally had tried to eliminate.
But a handful of nucleotides matter, too. Metabologenomics revealed that the presence or absence of two to three nucleotides — essentially letters that make up a genetic message— can tune the switches that drive PTM antibiotic production. This type of fine control previously had been found in certain bacteria that cause disease, but largely had been overlooked in bacteria that produce drugs.
“Our work highlights the problem of silent gene clusters and the need to understand them for next-generation drug discovery,” Blodgett said. “Comparative metabologenomics is a generally adoptable strategy, and we hope that others might use it to examine their own strains and drug pathways.”
Funding: This work was supported in part by the National Science Foundation under NSF-CAREER 1846005.