Nobody wants to be average.
But for a long time, scientists have found it convenient to think of bacterial cells as just that: “average.”
Researchers have traditionally relied on population-level strategies to understand fundamental aspects of bacterial physiology. These population-level approaches describe the behavior of idealized average cells, and they serve as the foundation for prevailing models of bacterial growth.
Models based on an average cell are useful, but they may not accurately describe how individual cells really work. New possibilities opened up with the advent of single-cell live imaging technologies. Now it is possible to peer into the lives of individual cells. In a new paper in PLOS Genetics, a team of biologists and physicists from Washington University in St. Louis and Purdue University used actual single-cell data to create an updated framework for understanding the relationship between cell growth, DNA replication and division in a bacterial system.
Petra Levin, the George William and Irene Koechig Freiberg Professor of Biology in Arts & Sciences at Washington University, an author of the new paper, has a keen interest in single-cell biology. In her research work, Levin has made seminal contributions to our understanding of bacterial cell growth.
A chance encounter at the Aspen Center for Physics led to a collaboration with Srividya Iyer-Biswas, a physicist at Purdue University with expertise in both first-principles-based physics theory and high-precision single cell experiments.
Taking advantage of the Zoom era brought on by the early days of the pandemic, Levin and Iyer-Biswas developed their virtual collaboration to revisit some of the “beautiful, classic models of the bacterial cell cycle,” as Levin describes them.
They found exciting bits were missing.

What was the problem? The models counted on the behavior of an “average” cell within a population. But using the average to infer what an actual cell does can be misleading.
“Imagine each bacterium as singing its own whimsical tune, following its own rhythm,” Iyer-Biswas said. “The collective — a population of millions of cells — has its own music, where no single voice especially stands out, but a song nonetheless emerges. From hearing just the collective rendition, how could one possibly uncover what precisely an individual’s song might be? That is the problem we were faced with.”
“What is true for the average cell is not necessarily true for the individual cell. Bacteria are just like us in this regard!” Levin added.
For this new paper, Levin and Iyer-Biswas worked together with Sara Sanders, a postdoctoral scientist in the Levin lab who recently moved to the National Institutes of Health (NIH), and Kunaal Joshi, a PhD student in the Iyer-Biswas lab, to tackle one basic question.
They wanted to figure out how these “whimsical” individual bacterial cells — or, as a more typical physicist might say, these stochastic cells — manage to exquisitely coordinate DNA replication with growth and division, so that overall events happen in the right sequence despite the “noisiness” of each process.
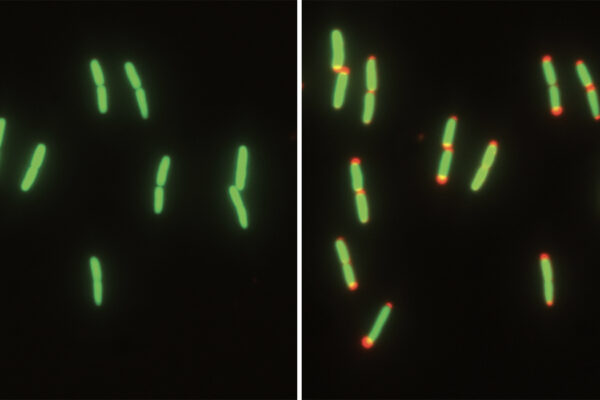
To answer the question, the authors carefully looked at single-cell growth data from the model organism Escherichia coli collected by the Jun laboratory at the University of California, San Diego. They then constructed a minimal mathematical model that captured complex, stochastic behaviors of individual cells and accurately matched individual cell data.
Based on average cell behavior, others had come to view the basic cell cycle steps of DNA replication and cell division as dependent on each other. But that wasn’t how Levin and Sanders saw it.
“Decades of genetic and molecular studies indicate that although DNA replication and division are clearly coordinated, they are not dependent on one another,” Levin said. “As long as there are mechanisms to prevent division across uncopied chromosomes, or fix the situation in the unlikely event that does happen, everything is fine. E. coli does not have cell cycle checkpoints like eukaryotic cells do.”
Meanwhile, Iyer-Biswas and Joshi realized that there was a simple way to understand the individual cell data. Each cell has three independent (stochastic) timers (equivalent to the whimsical tune from above) that start ticking each time DNA replication begins, and whose orchestration determines the sequence of cell cycle events.
Starting from this simple idea, Joshi discovered he could predict the sequence of DNA replication initiation, the end of DNA replication and division based on when the three timers independently go off and reset. His predictions matched exquisitely with the extant data on individual cell DNA replication and cell division in many different growth conditions.
By describing a stochastic, not deterministic, relationship between DNA replication and cell division, the authors have shifted how scientists understand a basic process in cell biology.
“Our ultimate goal is to build a community around high-precision approaches in biology that seamlessly integrate theory and experiment,” Iyer-Biswas said. “A more immediate goal is to transcend system-specific details and provide a unifying framework also applicable to other bacterial species.”