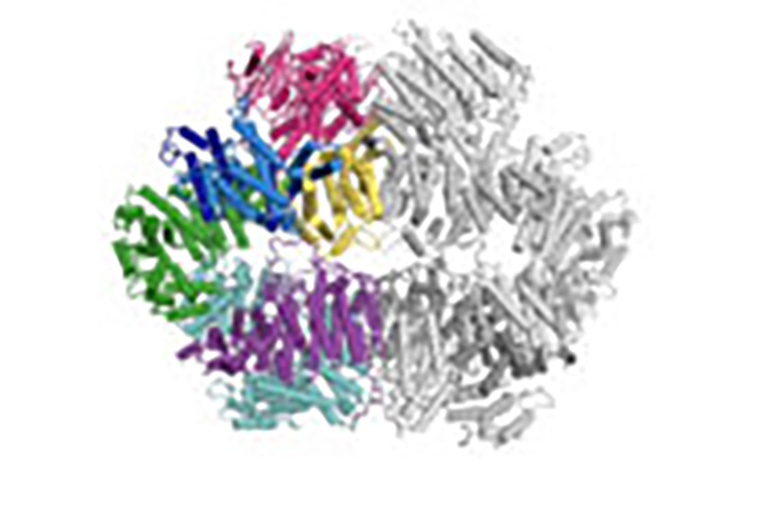
Researchers at Washington University School of Medicine in St. Louis and their colleagues at Imperial College London have identified how a key protein in cancer cells changes shape to kick-start the repair of DNA damage caused by chemotherapy or radiation. Blocking this built-in repair mechanism with a drug has the potential to make chemotherapy or radiation more effective, the researchers say. The key protein is called Mec1 in yeast and ATR in humans. The video shows two Mec1 molecules bound together (one is in color on the left; the other is grey on the right). The side in color shows how the protein moves to switch between active and inactive states. (Video: Luke Yates/Imperial College London) https://youtu.be/TUJiM92-i5U
Radiation and chemotherapy are designed to kill cancer cells. But for many patients, cancer cells can survive even after being hit with high doses of chemotherapy or radiation. To make treatment more effective, scientists are focusing on ways to tweak the inner machinery of cancer cells to make them more susceptible to dying.
A team at Washington University School of Medicine in St. Louis is making headway in such efforts. The researchers have identified how a key protein in cancer cells changes shape to kick-start the repair of DNA damage caused by chemotherapy or radiation. Blocking this built-in repair mechanism with a drug has the potential to make chemotherapy or radiation more effective, according to the scientists.
The study appears Nov. 9 in the journal Nature Structural & Molecular Biology.
Because this protein is essentially the same in lower organisms as well as people, the researchers studied the version of the protein found in yeast, called Mec1. Mec1 and its human counterpart, ATR, are activated when cells are stressed. The proteins are responsible for sensing and repairing DNA damage before cells replicate to prevent that damage from being passed down to daughter cells. In some cases, this activation is good, protecting healthy cells from DNA damage that could lead to cancer. But in other cases, such as cancer therapy, doctors would like to turn these repair mechanisms off so the cancer cells are more susceptible to death by further DNA damage. In this way, cancer cells — hit with radiation and chemotherapy — can be destroyed more easily.
“Determining the structures of both the inactive and active forms of this protein gives new insights into how that transition takes place, not just for Mec1 and ATR but for other members of the same family of proteins,” said senior author Peter M. Burgers, the M. A. Brennecke Professor of Biological Chemistry. “ATR inhibition is a promising anti-cancer treatment when combined with radiation or chemotherapy. A handful of ATR kinase inhibitors exists, and one called ceralasertib is being tested in phase 2 clinical trials in the U.S. Our study provides a tool for improving current ATR kinase inhibitors or designing new ones in a laboratory. Providing high-resolution structures is a critical step in the intelligent design of selective inhibitors.”
To determine these structures, the Burgers lab studied yeast with various mutations in this key protein and found one mutant that forced the protein into a permanent on position. With collaborators at Imperial College London, Luke Yates and Xiaodong Zhang, the researchers then determined the structure of the protein when constantly on, at an extremely high resolution — on the scale of individual atoms.
“We already knew what it looks like when it’s off,” said first author Elias A. Tannous, a senior scientist in the Burgers lab. “But there was a lot of speculation about what it looks like when it’s turned on. How does it change its shape? Does it break in two? Does it bind to something else? We didn’t know. And it was interesting to find that it changes shape like a butterfly opening its wings.
“These types of proteins control many aspects of the cell, from growth and viability to replication and response to stress,” Tannous said. “It’s the master machinery of DNA damage response — responsible for accurate DNA replication. If there is any error, it tells the cell to stop. This can be good or bad depending on the situation. In future research, we can use this knowledge of the structure to learn how to fine tune the activity of this type of protein, with the goal of using this information to design more effective cancer therapies.”