Slaying bacteria with their own weapons
By exploiting bacterial systems for iron scavenging, a WUSTL scientist hopes to rescue antibiotics rendered useless by resistance
The Centers for Disease Control and Prevention warned last fall that the U.S. faces “potentially catastrophic consequences” if it doesn’t act quickly to combat the growing threat of antibiotic-resistant infections, which kill about 23,000 Americans a year.
Timothy Wencewicz, PhD, assistant professor of chemistry in Arts & Science at Washington University in St. Louis, has an idea that might provide a solution to resistance that is broader and more effective than the invention of a new drug.
“Today when you walk in with the symptoms of a bacterial infection,” Wencewicz said, “you are treated with a broad-spectrum antibiotic because we lack the ability to identify a bacterial strain quickly. And that means every type of bacteria in your body is exposed to that antibiotic.” So even justified antibiotic use that follows medical protocols encourages resistance.
One solution, he said, is personalized antibiotic therapy. This would require both rapid bacterial identification and narrow-spectrum antibiotics. Tailored antibiotic therapy would not only extend the clinical lifetime of new antibiotics by better managing resistance, it might also revive old antibiotics that have been abandoned due to resistance, toxicity, or their inability to penetrate bacterial membranes.
Wencewicz is working on a drug delivery system that would target specific bacteria by exploiting small molecules called siderophores they secrete to scavenge for iron in their environment. Each bacterium has its own system of siderophores, which it pumps across its cell membrane before cleaving off the iron.
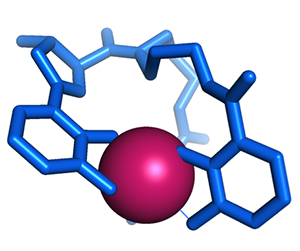
We counter the siderophore onslaught by making molecules called siderocalins that bind siderophores, preventing them from binding iron. But some bacteria sidestep this defense, making a sacrificial siderophore that saturates the available siderocalin and a second “stealth” siderophore that siderocalin doesn’t recognize.
“Each pathogen is unique,” Wencewicz said. “If we are going to use its own siderophores against it, we have to understand each siderophore it secretes, its function, and how its function is related to its structure.
“We have to quit thinking that every single bacterium is like all the other bacteria. They’re not,” he said.
Beware of Greeks bearing gifts
Wencewicz’ central insight is that this specificity—and the fact that the bacteria bring siderophores inside their cell membranes—can be turned against them.
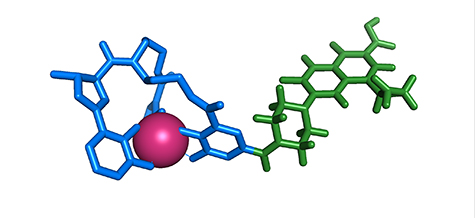
He is taking his cue from the bacteria themselves. Some bacteria, he said, link a toxic antibacterial agent to a siderophore, creating something called a sideromycin.
Competing bacteria, attempting to steal iron, grab the sideromycin and shuttle it in through their siderophore-uptake system. Once the sideromycin is inside the bacterium, the toxin it carries kills the bacterium.
Because the siderophore system is highly specific, Wencewicz believes it can be used to target even broad-spectrum antibiotics to individual bacterial strains, avoiding the indiscriminate exposures that spread resistance. And because the Trojan horse antibiotics will ferry antibiotic inside the membrane, only tiny doses will be needed.
Correnti, C.; Clifton, M. C.; Strong, R. K.; Raymond, K. N. ACS Chemical
Biology 2013, 8, 1882-1887)
For both reasons he believes siderophore delivery systems will delay the emergence of resistance to new antibiotics and rescue many older antibiotics that have been abandoned because of resistance or toxicity.
Targeted therapy would be useless without rapid diagnostics because doctors wouldn’t know which sideromycin-like drug to prescribe. But because siderophore systems are specific to bacterial species, they could be used to type as well as to treat bacteria.
Because of resistance, we really need to discover new approaches to antibiotic development and not just continually re-jigger existing drugs, Wencewicz said. The siderophore pathway offers the opportunity to completely re-imagine antibiotic development so that it takes resistance into account up front instead of on the back end.